Cyanobacteria are ubiquitous autotrophic bacteria that have the natural ability to utilize sunlight to convert atmospheric CO2 into carbohydrates. Cyanobacteria’s photosynthetic abilities are very appealing to synthetic biology and biotechnology.
Microfluidic chemostats provide an ideal environment in which the environmental conditions can be controlled tightly. The media can be exchanged easily during the experiment thus allowing for switching between for example carbon rich and carbon poor media or media with and without specific stress inducers.
The mother machine
design has the following advantages compared to other chemostats:
1. The mother cell can be followed for an unlimited period of time.
2. As the cells grow in a line, rather than randomly orientated like in the case of a normal
bacteria colony, image analysis (segmenting and single-cell tracking) is easier.
3. The media can still be easily exchanged.
Team Members:
Christian Schwall (cs687)*
Philipp Braeuninger-Weimer (pab96)
Bruno Martins (bmc36) *Primary contact
The Idea:
Cyanobacteria are ubiquitous autotrophic bacteria that have the natural ability to utilize sunlight to convert atmospheric CO2 into carbohydrates. Cyanobacteria’s photosynthetic abilities are very appealing to synthetic biology and biotechnology. Firstly, these organisms can be harnessed to produce compounds, such as biofuels, that they would not normally produce in the wild, but which have important economic value. Secondly, cyanobacteria possess a number of comparative advantages over land plants for this goal: their genomes are easier to manipulate, they have faster growth rates, and they can be grown in areas that are not suitable to agriculture (1).
While viable biotechnological applications of cyanobacteria will ultimately require production to take place in large reactors and tanks, the optimisation of the underlying cellular process will hinge on an appropriate quantitative understanding of how biochemical networks (both endogenous and synthetic) operate intracellularly. Metabolic and regulatory networks can interact with each other in non-intuitive ways, and often the mechanisms of such interactions are only apparent at the single cell level. However, while systems and synthetic biologists have spent considerable effort in observing and building tools, such as microfluidic chemostats, to study microbes at the single cell level, these efforts have largely focused on other types of bacteria. Organisms such as E. coli and B. subtilis have been preferred because of their ubiquity as laboratory model organisms and extremely fast growth rates.
Microfluidic chemostats provide an ideal environment in which the environmental conditions can be controlled tightly. The media can be exchanged easily during the experiment thus allowing for switching between for example carbon rich and carbon poor media or media with and without specific stress inducers.
A clever microfluidics design, known as the ‘mother machine’, grows bacteria in dead ended channels with a diameter of approximately one micron (growth channels) (2). The diameter of the channels is chosen to match the average diameter of an individual rod-shaped bacterium (Figure 1).
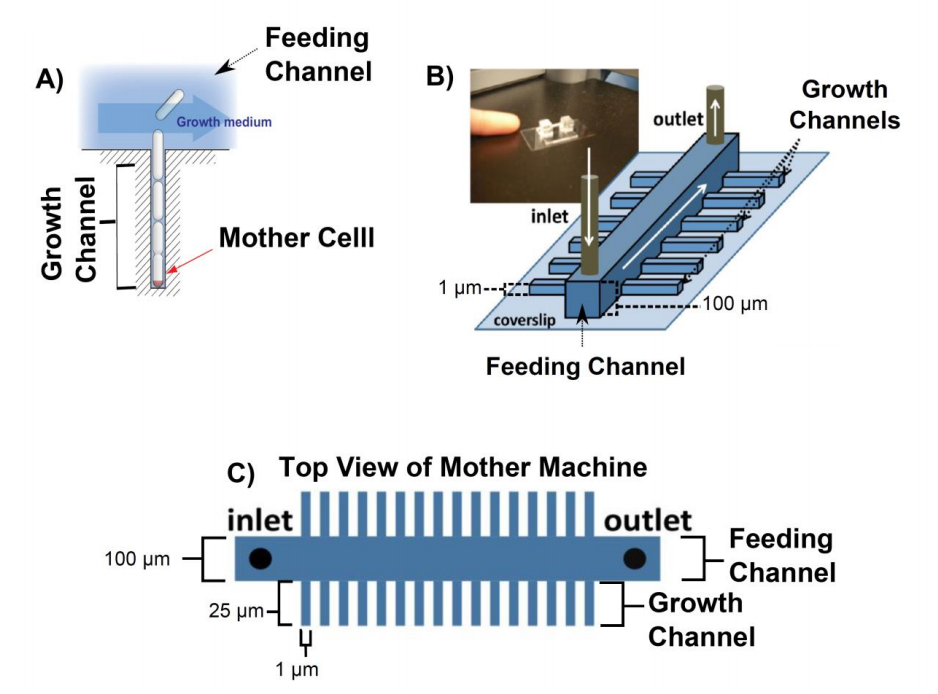
Figure 1. Overview of the mother machine. A) The mother cell is trapped at the bottom of a dead ended channel. As the cells divide they get pushed out of the growth channel into the feeding channel where they get flushed away. B) Sketch of the mother machine and a picture of the microfluidic device. C) Top view of mother machine. The large channel in the middle of the device is called feeding channel and the small channels branching out of the feeding channel are called growth channels. All figures were adapted from (1)
Due to this geometry, the cells grow in a line and the cell at the bottom of the dead ended growth channel (mother cell) can, in principal, be studied for an unlimited period of time (Figure 1 A). Fresh media is supplied through a comparably wide (100 µm x 100 µm, width x depth) feeding channel. Hundreds of short (25 µm in length) growth channels are attached to the feeding channel in a 90° angle. Any bacterium that is pushed out of the growth channel as the cells divide is flushed away into the feeding channel. Fresh nutrients reach the end of channel by diffusion. The mother machine design (2, 3) has the following advantages compared to other chemostats:
1. The mother cell can be followed for an unlimited period of time.
2. As the cells grow in a line, rather than randomly orientated like in the case of a normal bacteria colony, image analysis (segmenting and single-cell tracking) is easier.
3. The media can still be easily exchanged.
This device, and variations of it, are made out of polydimethylsiloxan (PDMS) and were fabricated from a patterned silicon wafer (master) using soft lithography (2–4). The masters themselves are fabricated by photolithography (2–4). The drawback of conventional photolithography is that its resolution limit is about 1 µm thus making the fabrication of micrometre sized growth channels very challenging. An alternative to photolithography in the fabrication of such small channels is electron beam lithography (EBL). Few devices have exploited EBL to fabricate mother machines due to the high cost of electron beam time and the EBL facilities compared to conventional photolithography. However the excellent control over the pattering of feature sizes down to 10 nm and the high success rate are of great advantage.
A mother machine that has been produced using EBL was presented by Moolman and co-workers. They patterned a positive photoresist with EBL and etched the channels into the silicon wafer (5) (Figure 2 A). As a result the pattern on the silicon wafer was not the negative of the final PDMS device and an intermediate mold made out of PDMS had to be fabricated (Figure 2 B) before the patterns could be transferred into PDMS (Figure 2 C).
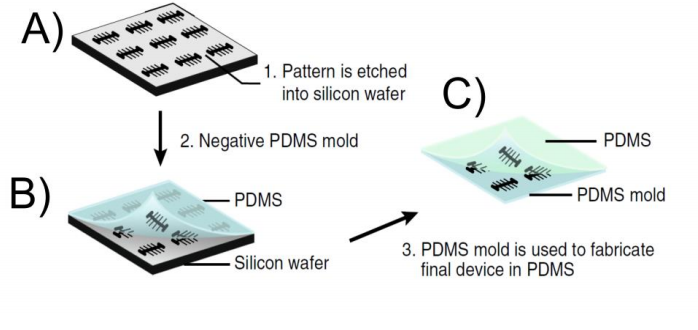
Figure 2. Workflow of Moolman device. A) The patterns are etched into the silicon wafer after the negative resist has been patterned with the electron beam. B) A negative replica of the structures on the silicon wafer is transferred into a PDMS mold. C) The PDMS mold made in B) is used to pattern the final device onto the PDMS. The final pattern on the PDMS is identical with the pattern on the silicon wafer. The figure was taken from (5)
In our project we will design a microfluidic device based on the mother machine, which will be optimized for the cyanobacterium S. elongatus PCC7942 and it will allow running experiments in multiple mother machines at the same time on one device for 7 days. Rod-shaped cyanobacteria, such as S. elongatus, can be loaded onto and grown in existing microfluidics devices (6, 7), but have a small number of specific requirements that are not always met by devices optimised for, e.g., E. coli and B. subtilis.
One important difference is S. elongatus cells differ in size, being typically longer and, more importantly, wider than other common types of rod-shaped bacteria (for example, and according to our own measurements, S. elongatus cells are 50% wider on average than B. subtilis cells). These cellular dimensions require matching channel dimensions for optimal experimental performance. Another important difference is cyanobacterial cells need a light source to proliferate. Under a typical widefield microscope, the light source is attached to the condenser lens and sheds light directly above the sample at very low illumination angles. Short distances between the light source and the sample, and low angles of incidence are problematic because it is then difficult to maintain even illumination across a modestly wide field of view.
Cyanobacteria grow an order of magnitude slower than E. coli and B. subtilis, and so, in order to speed up the experimental process, it would be convenient if one could test multiple strains and conditions in the same device, while keeping light conditions comparable. This goal can only therefore be achieved by building a microfluidics device with separate, but closely arranged channels that maximise space on the chip (Figure 4). To account for these requirements specific to cyanobacteria we divide the project into two main focus areas:
1. Using EBL to optimize the growth channel size to grow cyanobacteria
2. Optimization of the feeding channel layout on a device to assure even illumination on the device and increase the throughput.
Who we are:
Christian Schwall (cs687), Background: Physics/ Quantitative Biology
Philipp Braeuninger-Weimer (pab96), Background: Engineering
Bruno Martins (bmc36), Background: Physics/ Quantitative Biology
Implementation:
1. Using EBL to optimize the growth channel size to grow cyanobacteria
Our plan is to use EBL to fabricate growth channels specifically tuned to grow cyanobacteria similarly to Moolman and co-workers (5). However instead of using a positive resist we will be using a negative photoresist. Positive photoresists become soluble to the developer in the areas that have been exposed to the light (Figure 3 A). Negative resists are insoluble to the developer in the areas that have been exposed to the light (Figure 3 B).
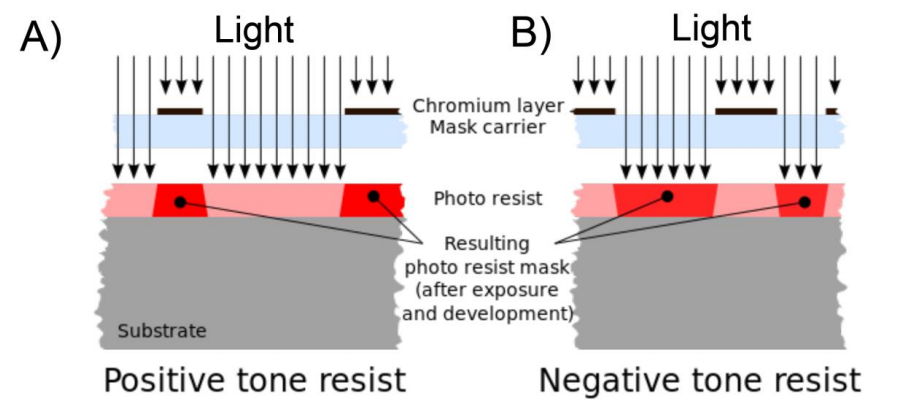
Figure 3. Overview of positive and negative resits. A) Positive photoresists become soluble to the developer in the areas that have been exposed to the light. B) Negative resists are insoluble to the developer in the areas that have been exposed to the light. The image was taken from Wikipedia (8).
The advantage of negative resists is that the patterns can be easily transferred into PDMS as the pattern on the silicon wafer is the negative of the final device in PDMS. Thus by using a negative resists with the EBL the intermediate PDMS mold Moolman and co-worker needed for the fabrication of their device (Step 2 in Figure 2) would no longer be necessary. This would greatly facilitate the replica molding of the patterns from the silicon wafer to the PDMS. Unfortunately, the choice of negative EBL resists is very limited and a new process flow has to be developed. We have narrowed down our choice of negative resists to AZ 5214E and SU8 (one of the most commonly used negative photoresists). Once we have found a good photoresist we will test a variety of growth channel sizes to optimize the growth conditions of cyanobacteria in our mother machine.
2. Optimization of the feeding channel layout
On a typical mother machine there is only one feeding channel, which allows for one experiment at a time (Figure 4 A). This greatly limits the application of the mother machine to slow growing cyanobacteria. We therefore want to design a device with multiple mother machines packed closely to each other such that they can fit onto a chip of the size of a 2 pound coin (Figure 4).
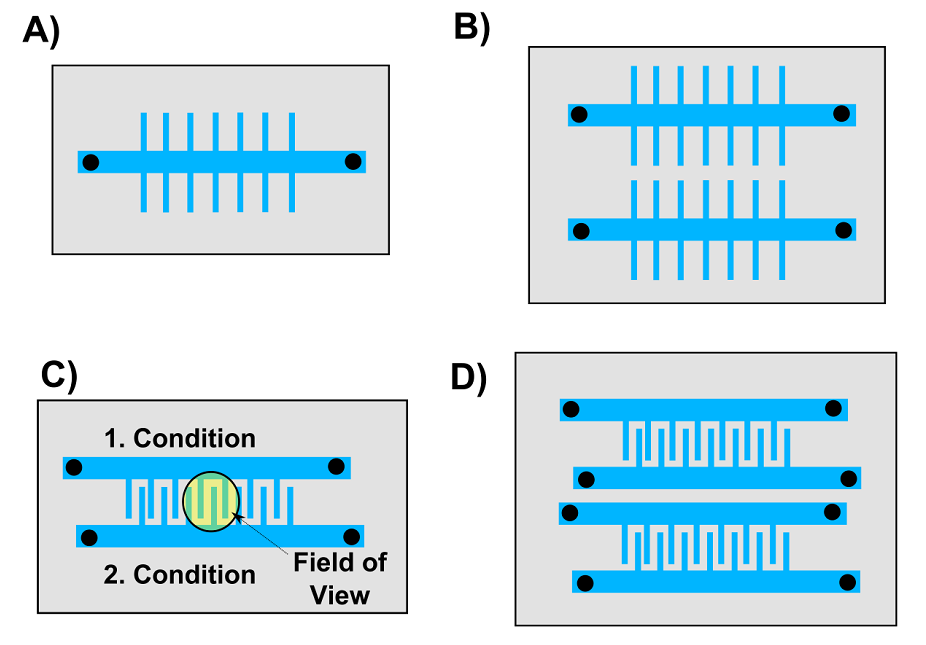
Figure 4. Overview of mother machine designs we want to test. A) The classic mother machine device published by Wang and co-workers. B) This device has two mother machines on one chip. C) In this device two growth channels with different conditions and strains can be image in one field of view (radius = 600 µm). D) A combination of B) and C).
To test the feasibility of our project we fabricated a mother machine entirely by using EBL to pattern channels of a constant depth of 1 µm and a width of 1 µm to 10 µm into AZ 5214E. The structures were replicable multiple times into PDMS without losing any of their resolution and we were able to load cells into the growth channels (Figure 5). Unfortunately we could not establish a good perfusion of media through the feeding channels as they were too shallow. Thus, we now want to fabricate feeding channels with a width of 100 µm and a depth of 100 µm as Wang and co-workers did (2). Pattering such large structures using EBL is tedious which is why we will exploit conventional photolithography in a second step before the growth channels have been deposited. For the photolithography step we will have to order various chrome masks to pattern the photoresists. These chrome masks are expensive and unfortunately there is no alternative as transparencies or emulsion glass masks do not have a high enough resolution.
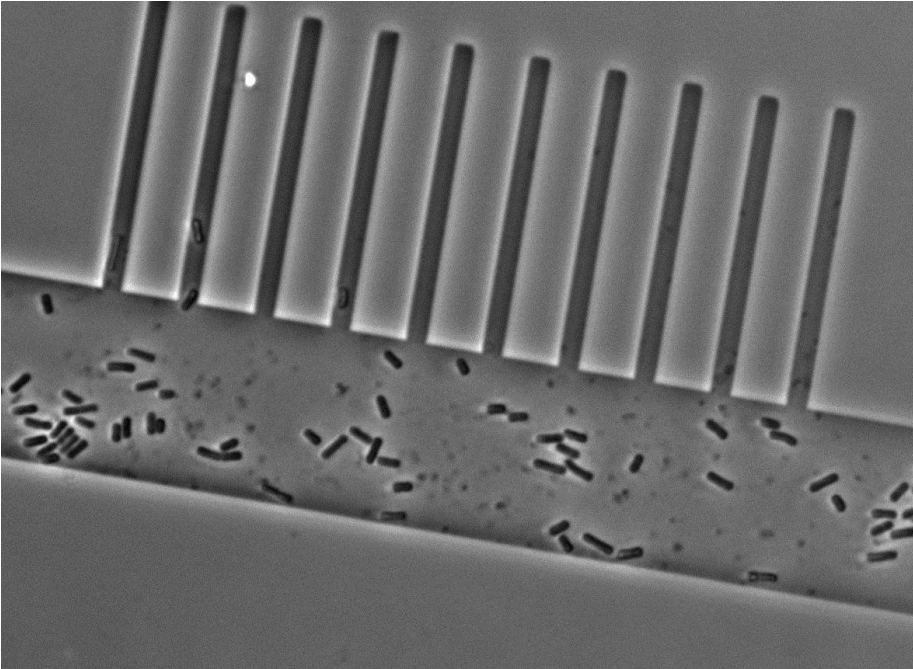
Figure 5. Micrograph of cells in the prototype mother machine.
The work in the project will be divided as follows: - All the necessary clean room work e.g. the lithography work will be done by Philipp Braeuninger-Weimer who is an expert on EBL and photolithography. - The design of the microfluidic chips and the PDMS work will be done by Christian Schwall. - The devices will be tested with cyanobacteria by Christian Schwall and Bruno Martins.
Benefits and outcomes:
To check the feasibility of our project we made a promising prototype fabricated with electron beam lithography (EBL) in collaboration between the Department of Engineering (Philipp Braeuninger-Weimer) and the Sainsbury Laboratory (Christian Schwall and Bruno Martins). The funding for electron-beam time and access to the clean room facilities is already in place. In order to further pursue this project, what we require at this point is funding to design a few chrome masks so we can pattern the larger feeding channels using conventional photolithography. The new device would be specifically designed for cyanobacterium S.elongatus PCC 7942, which to our knowledge, is a device that does not yet exist. Further, having multiple mother machines very close to each other on the same chip would drastically increase the throughput of one experiment. This is of great importance when running experiments for 7 or more days.
The direct benefits of this project to the framework of synthetic biology are two-fold:
1. S. elongatus is one of the most widely studied cyanobacteria, because it is the model organism of choice for research on the prokaryotic circadian clock, and because it is a promising host of synthetic circuits for metabolic engineering (1).Despite their importance, specific single-cell analysis tools have, so far, prioritized other types of bacteria. We believe that, in order to optimally characterise and manipulate the interaction between a cyanobacterium ‘chassis’ (i.e., a cell) and an exogenous circuit, we must first study systems in a rigorously controlled environment at the single-cell level. Our device will provide such a level of control. On the other hand, while we will design and test our device with cyanobacteria, there is no impediment to it being tweaked and used to grow other bacteria.
2. Microfluidics has emerged as a pivotal technique to study organisms at single-cell or microcolony levels. However, fabricating the devices can be challenging and discourage usage by the wider community. It is therefore essential that cross-talks between advanced engineering and fundamental biology are maintained. Our proposal addresses this cross-talk by: i) partnering skills in materials engineering, physics and cell biology from two different Cambridge departments; ii) exploring the potential of EBL, a technology that has not yet been used extensively to fabricate devices for biology; iii) accomplishing all steps that link an engineering blue print to a real everyday experiment in a biology lab.
Other researchers will benefit from our project too: for any working design, we will share the CAD files and our gained expertise openly. We will produce a step-by-step manual, or equivalent documentation, which will be shared online. Anyone in the community will be able to use or modify our device for their own needs.
Budget:
We budget our project as follows:
1. Alignment Mask: £ 600.00
2. Mask for device with one feeding channel per chip (Figure 4 A): £ 600.00
3. Mask for device with multiple feeding channels per chip (Figure 4 B): £ 600.00
4. Mask for device with two feeding channels per chip and two growth channels per field of view (Figure 4 C): £ 600.00
5. Mask for device with multiple feeding channels per chip and two growth channel per field of view (Figure 4 D): £ 600.00
6. Spare Mask: £ 600.00 7. Consumables: £ 400.00
Total: £ 4000.00
Additional support for the project will be provided as follows:
- Free access to the clean room and electron beam time will be provided by Philipp Braeuninger-Weimer.
- Dr James Locke as agreed to make his soft lithography lab (plasma cleaner, oven and desiccator) and cyanobacteria strains available for project.
- Additional funding for consumables will be provided by Dr. James Locke
References:
- I. M. P. Machado, S. Atsumi, Cyanobacterial biofuel production. J. Biotechnol. 162, 50–56 (2012).
- P. Wang et al., Robust growth of Escherichia coli. Curr. Biol. 20, 1099–1103 (2010).
- T. M. Norman, N. D. Lord, J. Paulsson, R. Losick, Memory and modularity in cell-fate decision making. Nature. 503, 481–6 (2013).
- Z. Long et al., Microfluidic chemostat for measuring single cell dynamics in bacteria. Lab Chip. 13, 947–54 (2013).
- M. C. Moolman, Z. Huang, S. T. Krishnan, J. W. J. Kerssemakers, Electron beam fabrication of a microfluidic device for studying submicron-scale bacteria. J. Nanobiotechnology. 11, 12 (2013).
- J. R. Moffitt, J. B. Lee, P. Cluzel, The single-cell chemostat: an agarose-based, microfluidic device for high-throughput, single-cell studies of bacteria and bacterial communities. Lab Chip. 12, 1487 (2012).
- S.-W. Teng, S. Mukherji, J. R. Moffitt, S. de Buyl, E. K. O’Shea, Robust Circadian Oscillations in Growing Cyanobacteria Require Transcriptional Feedback. Science (80-. ). 340, 737–740 (2013).
- Cepheiden, Comparison positive negative tone resist. Wikipedia, (available at http://en.wikipedia.org/wiki/Photoresist).

